Stereopy - Spatial Transcriptomics Analysis in Python#
Stereopy is a fundamental and comprehensive tool for mining and visualization based on spatial transcriptomics data, such as Stereo-seq (spatial enhanced resolution omics sequencing) data. More analysis will be added here, either from other popular tools or developed by ourselves, to meet diverse requirements. Meanwhile, we are still working on the improvement of performance and calculation efficiency.
Get quickly started by browsing Usage Principles, Tutorials or API.
Open to discuss and provide feedback on Github.
Follow changes in Release Notes.
News#
The paper of Stereopy has been pre-printed on bioRxiv!
Upcoming functions#
Batch Effect removal funciton
Lasso expression matrix and image simultaneously
…
Highlights#
More suitable for performing downstream analysis of Stereo-seq data.
Support efficient reading and writing (IO), pre-processing, and standardization of multiple spatial transcriptomics data formats.
Self-developed Gaussian smoothing model, tissue and cell segmentation algorithm models, and cell correction algorithm.
Integrate various functions of dimensionality reduction, spatiotemporal clustering, cell clustering, spatial expression pattern analysis, etc.
Develop interactive visualization functions based on features of Stereo-seq workflow.
Workflow#
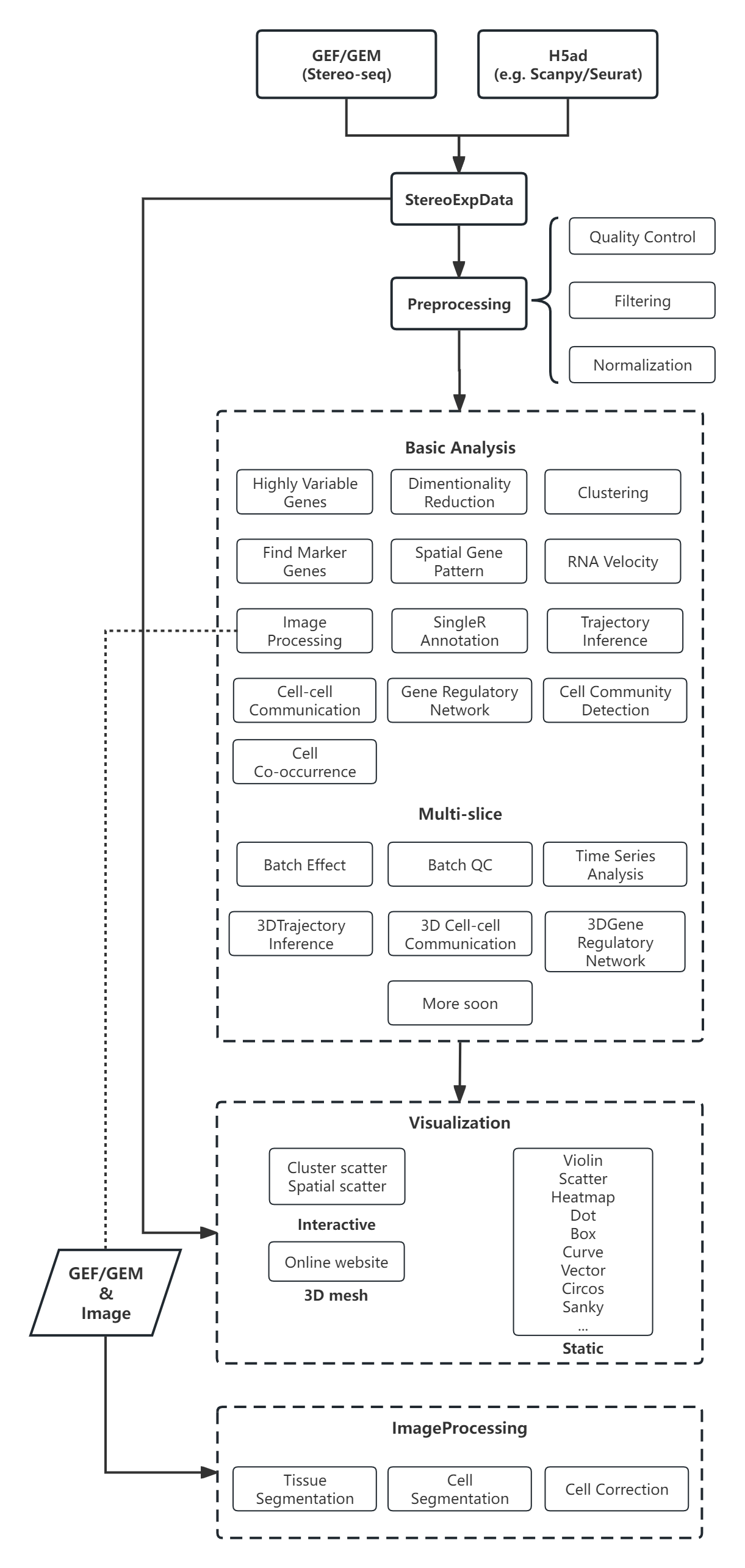
Latest Additions#
Version 1.2.0#
1.2.0 : 2024-03-30
Features:
st.io.read_gemandst.io.read_gefsupport expression matrix files with geneID information.Analysis results of
find_marker_geneswill be saved into the output AnnData h5ad.Upgraded tissue segmentation algorithm.
Addition of
st.tl.adjusted_rand_scoreto calculate the adjusted Rand coefficient between two clusters.Addition of
st.tl.silhouette_scoreto calculate the average silhouette coefficient of a cluster.h5ad2rds.Ris compatible with AnnData version > 0.7.5, to convert from h5ad to rds files.Addition of the clustering category labels to the graph of
st.plt.paga_compare.
BUG Fixes:
Fixed the error of high memory consumption when converting
X.rawinto AnnData.
Version 1.1.0#
1.1.0 : 2024-01-17
Features:
Reconstructed
st.plt.violinvisualizing function which is now not only applied to display QC indicators;ins.export_high_res_areacan handle expression matrix and image simultaneously, to lasso region of interest and corresponding sub-image.Interactive visualizing
st.plt.cells_plottingsupported displaying expression heatmap and spatial distribution of a single gene.When input GEF and GEM at cell level, information of DNB count and cell area would be added into
cells/obs, and cell border would be added intocells_matrix/obsm.
BUG Fixes:
slideiopackage removed historical versions, resulting in an installation failure.Calculating error when performing
ms_data.tl.batch_qc, due to abnormalos.getlogin.st.plt.paga_time_series_plotindicated that the image was too large to draw, due to unprocessed boundary values when computing median.
Version 1.0.0#
1.0.0 : 2023-12-04
Features:
Addition of GPU acceleration on SinlgeR for large-volume data, and optimized calculating based on CPU version.
Addition of
st.plt.elbowto visualize PCA result, for appropriate number of pcs.Addition of color, max, min setting for colorbar, when plotting heatmap.
Addition of cell segmentation of
Deep Learning Model V1_Pro, which is improved based onV1.Supplemented parameters of
st.plt.auc_heatmapandst.plt.auc_heatmap_by_group, full access toseaborn.clustermap;Addition of thread and seed setting in
st.tl.umap, of which the default method have been changed to single thread with the sacrifice of computational efficiency to ensure reproducibility of results. More in https://umap-learn.readthedocs.io/en/latest/reproducibility.html.Modification of computing method of bin coordinates when reading GEM, consistent with GEF.
Optimized
st.io.stereo_to_anndatafor efficient format conversion.Renamed
st.tl.spatial_alignmentfunction asst.tl.paste.export_high_res_arearemoved parametercgef.
BUG Fixes:
Occasional square-hollowing area in
Deep Learning Model V3of cell segmentation processing.st.tl.annotationcould not set two or more clusters as a same name.The data object
ins.selected_exp_dataobtained fromst.plt.interact_spatial_scattercould not be used for subsequent analysis.Part of data was missing when performed
st.plt.interact_spatial_scatterto output high-resolution matrix in GEF format.Some files met reading error, led by no default setting of
bin_typeandbin_sizeinst.io.read_h5ms.Error in Batch QC calculation due to data type problem.
There is NaN in Cell Community Detection output after threshold filtering, resulting in a calculating error when performed Find marker genes based on it.
st.plt.paga_time_series_plotindicated the image is too large to draw, leading to graph overlap, due to the limitation of matplotlib package.



